|
Research
The
Systematic Bioengineering Laboratory (SBL) at Penn State develops precision bioengineering
technologies, such as single
cell biosensors, microfluidic
systems, and data science and AI workflows, for precision
medicine applications. These technologies have
been applied to a variety of biomedical applications, including antifouling
coating for sanitation and medical applications (Nature Sustainability 2019),
point of care diagnostics of urinary stone disease (Science Advances 2020),
and epigenetic priming of iPSC reprogramming (Nature Materials 2022). Currently,
our focus is on leveraging these technologies to investigate regulatory
mechanisms of collective
cancer invasion, establish rapid diagnostic systems for infectious diseases and dysbiosis, and develop personalized
immunotherapies.
Technological Development
Biosensor Design for Dynamic Single
Cell Analysis. SBL has made significant contributions to biosensing
in live single cells for dynamic multigene analysis. For instance, we have
established GNR-LNA biosensors for mapping dynamic gene expression profiles
in photothermally stimulated lung tissues, mechanically damaged mouse
cornea, Nrf2 mediated chemoresistance in KRASG12D mouse lung tumors, and
patient-derived tumor organoids (ACS Nano 2014 Link; Advanced Materials 2015 Link). In addition, we
have demonstrated multiplex detection of both mRNA and protein in the same
cell by incorporating molecular aptamers into the biosensor design (Biomaterials 2018 Link). These single
cell biosensors have been applied to rapidly identify bacterial species at
the single cell level (Nanomedicine: NBM 2019 Link) and to monitor the
reprogramming of epigenetically primed iPSCs (Nature Materials 2022 Link).
|

GNR-LNA biosensors
|
Bioinspired microfluidics for 3D
tissue modeling and disease diagnostics. SBL has pioneered
microfluidic devices and single cell manipulation techniques for medical
diagnostics and tissue modeling. For instance, we confine pathogens from
raw or enriched patient samples in microchannels to determine the bacterial
antibiotic resistance profiles at the single cell level (PNAS 2019 Link). Their growth
rates and antibiotic resistance profiles can be determined at the single
cell level in as few as 30 min. We have also developed bioinspired microfluidic systems for the metabolic
evaluation of urinary stone disease at the point of care (Science Advances 2020 Link). Our technologies
are being adopted in various clinical studies and product development pipelines
worldwide, in partnership with clinical and industrial partners (Nature Sustainability 2019 Link).
|

Single cell AST device
|
Data science and artificial
intelligence. SBL is engaged in data science and computational
techniques (PLoS Computational
Biology 2016 Link). For instance, we
establish an artificial intelligence (AI)-guided experimental strategy for
screening potent antiviral drug combinations and immunomodulation cocktails
(PNAS 2008 Link). The AI-guided
method reduces the one million possible cases in the search space into as
few as ten iterations, which dramatically reduces the time and cost of the
optimization process. Similarly, we have demonstrated a metamodel
antimicrobial cocktail optimization (MACO) scheme to identify synergistic
antibiotic cocktails that reduce the minimum inhibitory concentration
40-fold (PLoS ONE 2010 Link). We are actively working on machine learning and
bioinformatic techniques for modeling complex biomedical processes and
medical diagnostics.
|
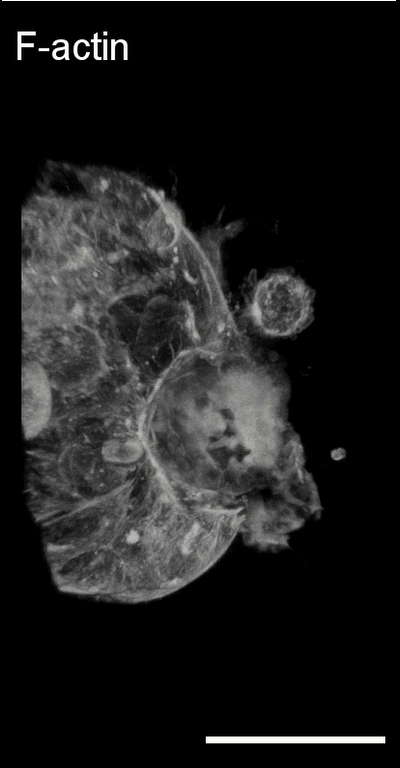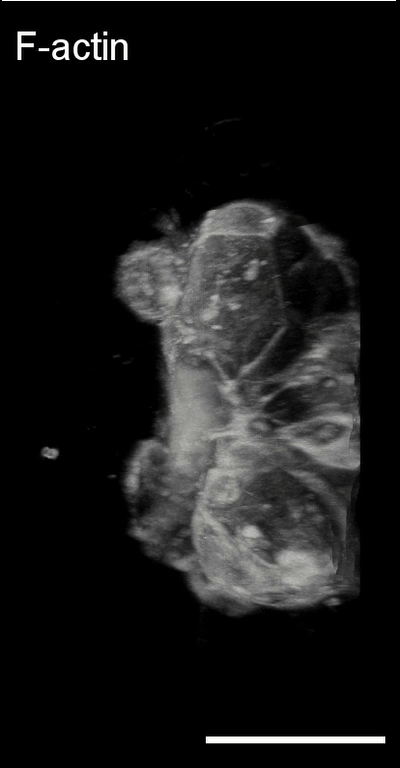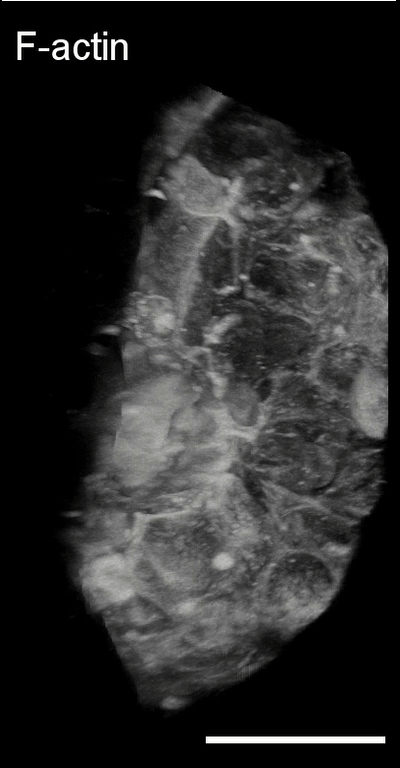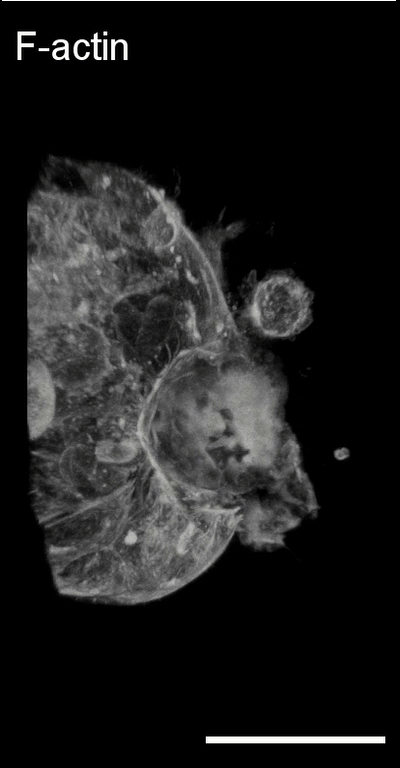
3D image analysis
|
Biomedical
Applications
Infectious diseases and antimicrobial
resistance. Rapid detection of pathogenic agents is critical
toward the judicious management of infectious diseases, such as urinary
tract infection and sepsis, especially in emergency situations and
high-risk areas such as hospitals, airports, rural clinics, and temporary
clinics established in response to disasters (Nature Biomedical
Engineering 2020 Link). In settings
where highly infectious pathogens are suspected, point-of-care detection
will lead to the timely initiation of appropriate treatments, which will
reduce the infected individuals’ morbidity and mortality, as well as
address public health concerns by efficient triaging of the uninfected from
the infected. Within this context, we design and implement microfluidic,
rapid diagnostic systems to address the unmet critical need for rapid
pathogen identification and antimicrobial susceptibility testing (PNAS 2019 Link).
|

Single cell pathogen
identification
|
Collective cancer invasion and leader
cell formation. Collective cancer
invasion is increasingly recognized as a predominant mechanism in the metastatic
cascade. At the onset of collective cell migration, a subset of cells
within an initially homogenous population acquires a distinct “leader”
phenotype (Nature
Reviews Cancer, 2020 Link). However, the
molecular mechanisms driving the formation of invasive leader cells, as
well as the signaling network regulating their density during collective
cancer invasion, remain to be determined. Using dynamic single cell gene
expression analysis and computational modeling, we have shown that the
leader cell identity is dynamically regulated by Dll4-Notch1 signaling and
intercellular tension (Nature Communications 2015 Link). Furthermore, we
have elucidated the relationship between Nrf2, hybrid epithelial-mesenchymal
transition, and Notch1-Dll4 signaling in the regulation of collective cancer
invasion (PNAS, 2023 Link).
|
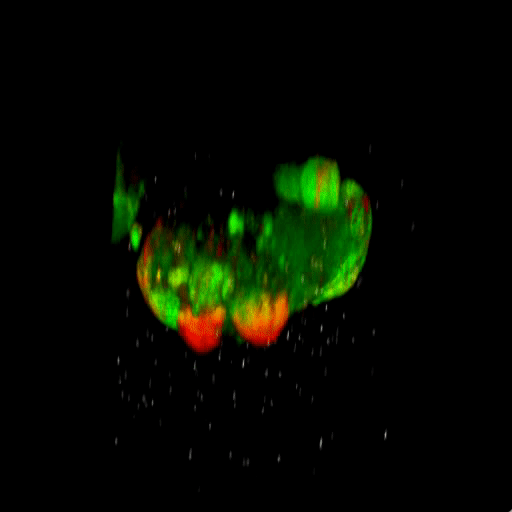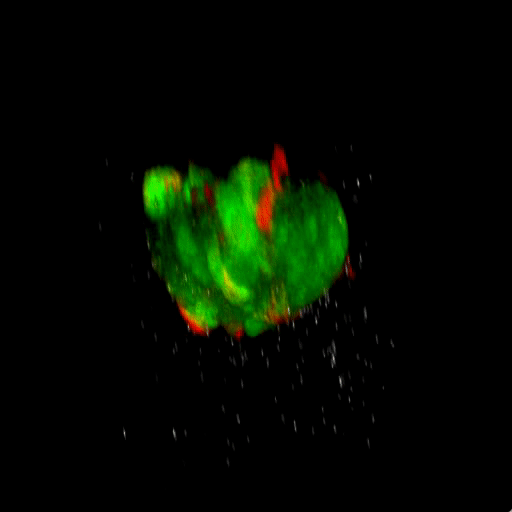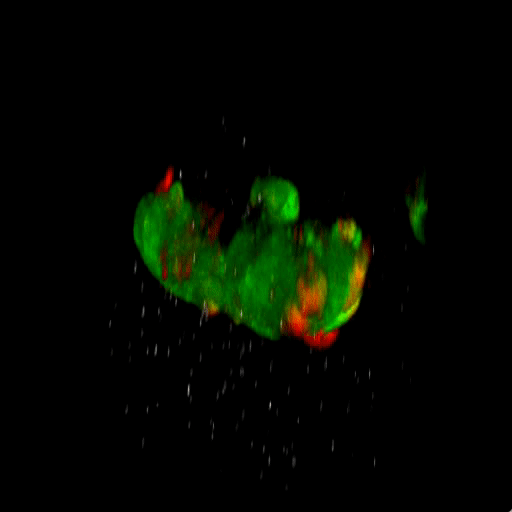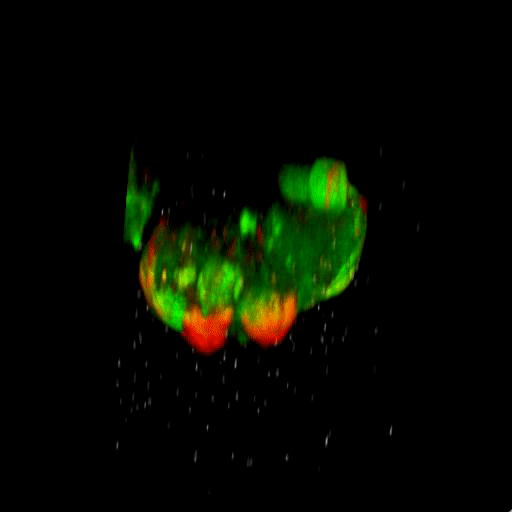
Bladder tumor organoids
|
Microbiota and immunotherapy. Microbiota
contribute fundamentally to human health, and the imbalance of microbiota
(dysbiosis) is associated with various medical conditions. Patients in the
hospital (e.g., ICU) are certain to experience disturbances of the
microbiota due to underlying diagnosis at admission and unintended
consequences of medical treatment. Furthermore, the microbiota is
increasingly recognized as a critical component in cancer and cancer
therapy (e.g., immunotherapy). Understanding the microbiota in various
medical conditions could open new opportunities for disease diagnostics, prognosis,
and treatment (Nature
Reviews Bioengineering; SLAS Technology 2019 Link).
|
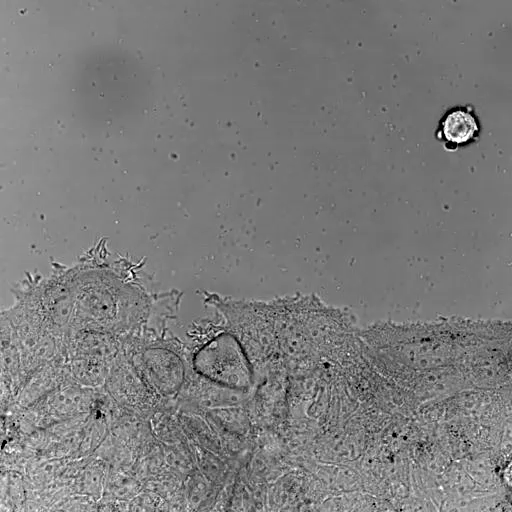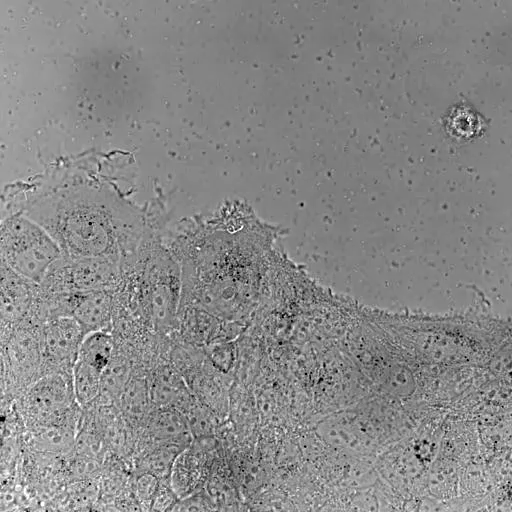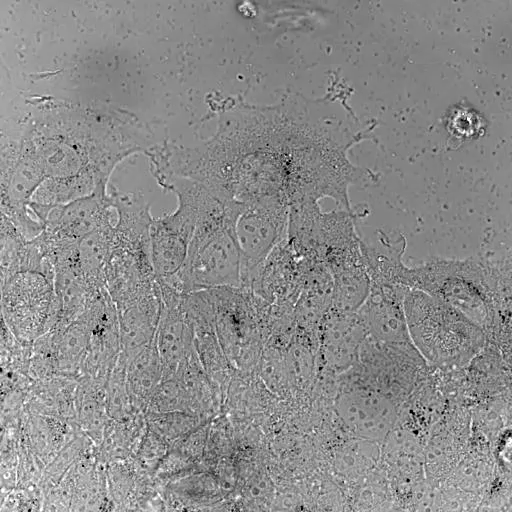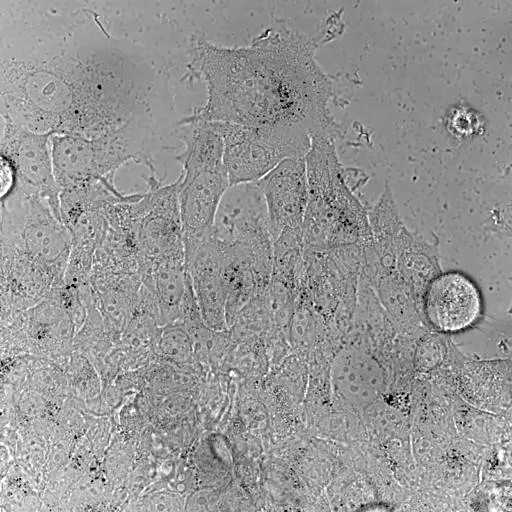
Single cell analysis
|
|






